Data Import
- You can now import 1H and 13C NMR data, structures, and assignments from third-party generated JCAMP-DX format files into the prediction database
- You can now import data from the following formats:
- MS data from Bruker TIMS TOF (*.tsf)
- GC data from Shimadzu (*.gcd)
- We improved data import from:
- Standard BMRB bioNMR (*.seq and *.bmrb) biosequence structure and assignment data is now transferred to C-H correlation spectra (HSQC and HMQC), in addition to the previous N-H HSQC
Auto Scroll to Create New Projects in a Full Projects Pane
- You can now auto scroll to Create new project at the bottom of a full Projects pane when dragging and dropping spectra
Updated Prediction Databases
- You can now predict spectra based on databases with thousands more structures, chemical shifts, and coupling constants than ever before
|
Structures (difference from previous update) |
Chemical Shifts (difference from previous update) |
Coupling Constants (difference from previous update) |
| 13C + 1H NMR Database |
392,075 (+8,095) |
5,609,600 (+219,550) |
1,152,584 (+53,401) |
| 13C NMR Database |
262,052 (+7,580) |
3,741,520 (+154,172) |
152,558 (+9,153) |
| 1H NMR Database |
283,215 (+7,969) |
2,768,019 (+123,582) |
1,044,497 (+44,975) |
| 19F NMR Database |
36,038 (+4101) |
62,402 (+5,429) |
89,264 (+12,996) |
| 15N NMR Database |
10,362 (+34) |
24,657 (+97) |
5,087 |
| 31P NMR Database |
33,216 (+782) |
40,250 (+836) |
51,484 (+2,108) |
New Integral Normalization Method for NMR Spectra
- You can now more accurately normalize 1H integrals of samples with various impurities with the Allow Excess option
- This can help prevent false negatives in workflows like Automated Structure Verification (ASV)
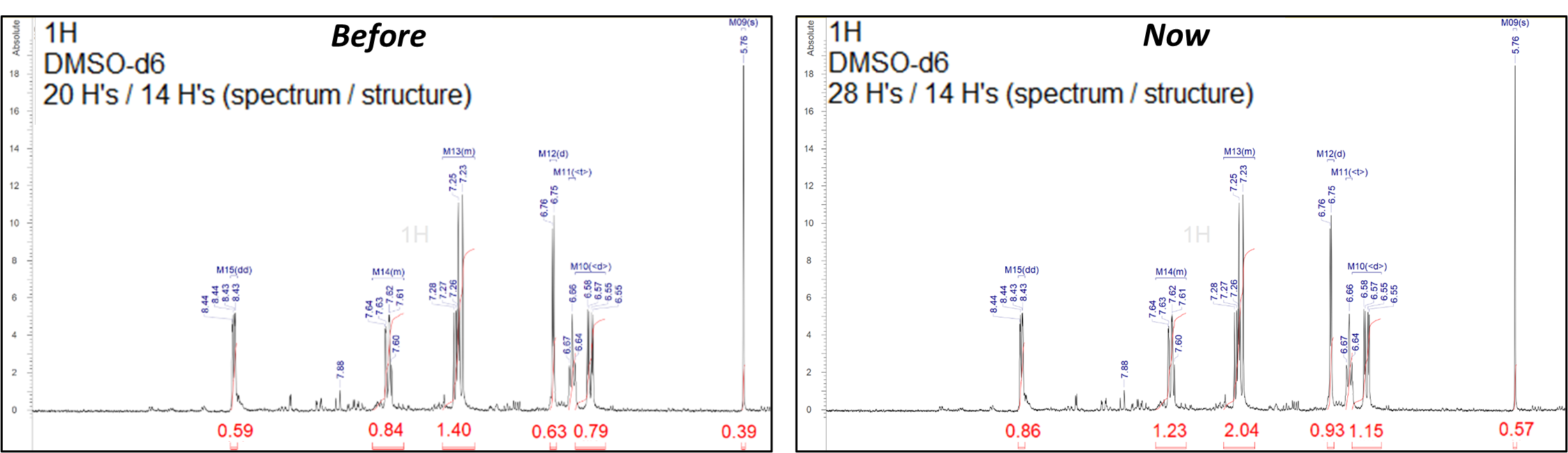
The Allow Excess option method for 1H integral normalization can avoid under-estimated integrals when impurities are present in the sample, which helps prevent false negative results in ASV.
Perform External Standard Concentration Calculations Without a Sample Structure
- You now have the option to perform qNMR external concentration calculations in cases where the structure of the sample is not available. Do this by entering the molecular weight of the sample in the Concentration dialog box.
Improved Metrics for External Standard qNMR
- You can now calculate the relative sample purity by weight (w/w%) automatically with the Concentration Calculation tool
- You can now include the concentration calibration parameters in your report
Option to Show Only Unique J-Coupling Constants in the NMR Spectral Data Table
- You now have the option to show only unique coupling constants in the Spectral Data Table Advanced Options menu
- Instead of reporting a triplet as t(5.25, 5.25), it can now be reported as t(5.25)
As always, you can process data from other analytical techniques in NMR Workbook Suite. We’ve improved features for these techniques as well:
Processing Hyphenated MS Data with NMR Workbook Suite
Improved Display and Reporting of Structures on Chromatographic Traces
- You now have the option to selectively display structures on chromatographic traces, consolidating information in one place. This feature allows you to control which structures to display and include in reports.
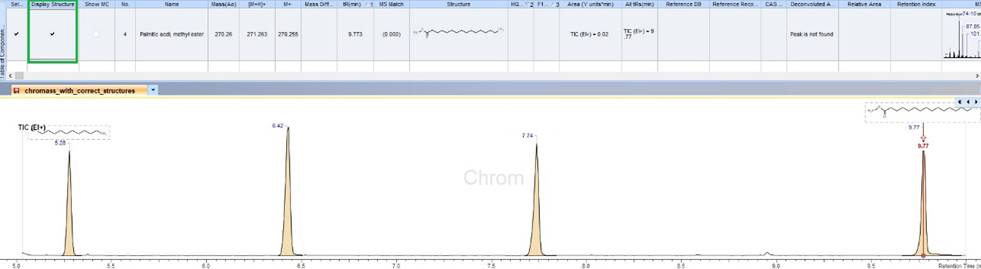
Display selected structure(s) on chromatographic traces.
Simplified Analysis with Reoccurring Ions
- You can now add reoccurring ions to the Component Interpretation Options, allowing you to automatically search and label reoccurring ions when assigning a component
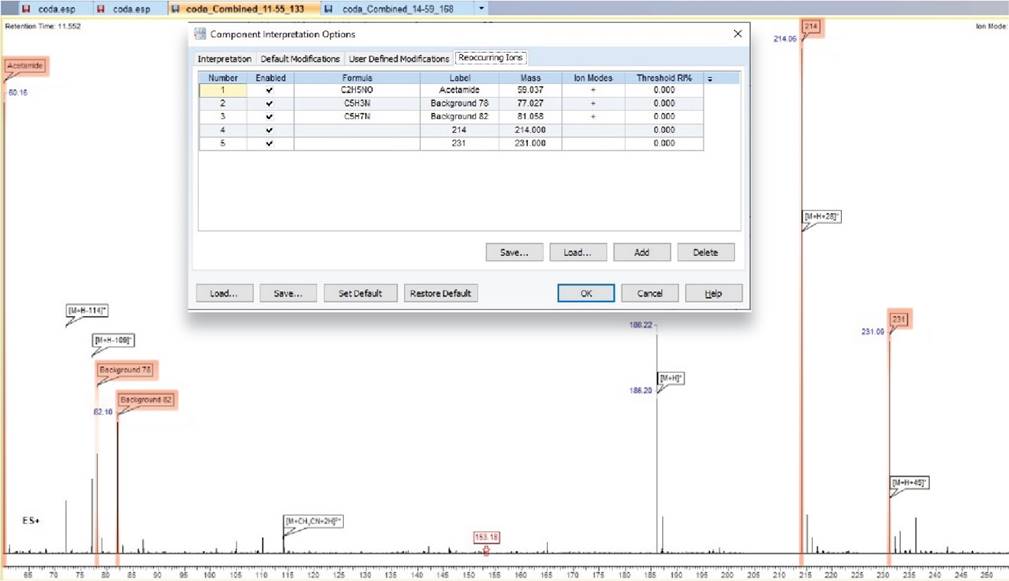
Highlighted ions are those populated in the reoccurring ions tab and are clearly labeled on the spectrum for ease of interpretation.
Greater Confidence in Identification with Improved MS Match Score
- You can now include multiply-charged ions and/or modifications into your MS Match score, giving you greater confidence in identification
Improvements in the Use of Internal Standards in Hyphenated Data Sets (LC/UV/MS, GC/MS)
- You can now calculate relative areas for all components if one component is selected as the internal standard
Easier Export of Components to Excel
- You now have the option to selectively push results including structure and molecular formulae from the Table of Components to Excel
Improved Accuracy in Area Calculations
- Using improved peak picking for XICs you can now manually adjust the border and/or baseline of peaks on XIC traces
Improved Efficiency of MS Data Processing
- You can now use preset options for Profiles to link instruments to specific accuracy, reporting, preferences, and script settings
- Set known parameters by instrument to minimize errors and improve efficiency
Improved Visualization of MS and UV Spectra
- You can now more easily review data with fixed positions for UV and MS Spectra
- Component Spectra are permanently fixed on the left and scan spectra are on the right
Improved Regulatory Compliance
You can easily store and visualize XIC accuracies and identify the accuracy used to extract XICs.
- Optionally display accuracies in the LC/UV/MS legend, in the chromatogram legend, and appended to the chromatogram name
- Visualize any accuracy changes in the History Record
- Store and display XIC accuracy in Spectrus DB, and visualize in reports
Improved Reporting Functionality
We have added more options for reporting functionality including:
- Ability to include full and deconvoluted areas in reports
- Ability to zoom to a defined region around an MSn Precursor
- Report “All” wavelengths for flat UV traces inside hyphenated datasets
- Display multiple descriptors on peaks in flat chromatograms such as retention time and name
Ease of Use Improvements
- You can access all available options in mass spectral right-click menus at once
- Quickly restore your zoom with a new hotkey for zoom undo (“U”)
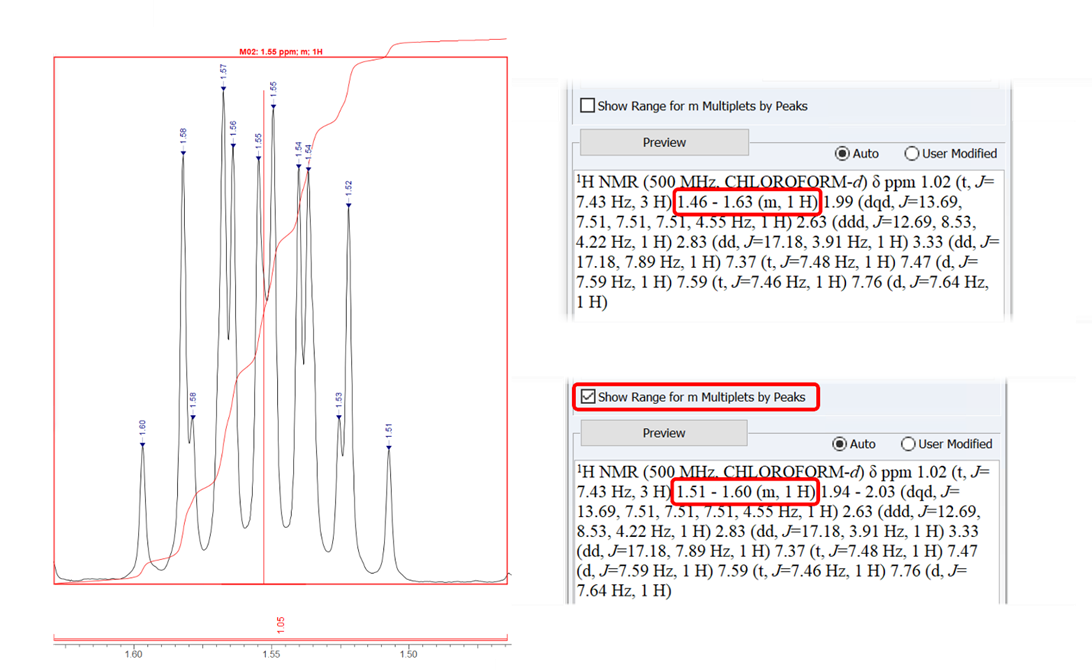 Choose to report multiplet ranges according to the outermost peaks or the boundaries of the integration—either 1.51–1.60 (m, 1H) for the outermost peaks with the option checked (above), or 1.46–1.63 (m, 1H) according to integration boundary (below).
Choose to report multiplet ranges according to the outermost peaks or the boundaries of the integration—either 1.51–1.60 (m, 1H) for the outermost peaks with the option checked (above), or 1.46–1.63 (m, 1H) according to integration boundary (below).