Version 2024 of ADME Suite introduces significant improvements to the P-gp Substrate specificity predictions including a new quantitative efflux model, expansion of the training libraries for P-gp Substrates and Protein Binding. Read below for details, and contact us for help upgrading your software.
New P-gp Efflux Ratio Module in the Quantitative PhysChem Model
This release implements a dedicated P-gp Efflux Ratio module, comprised of an advanced mechanistic physicochemical model based on Gradient Boosting statistical approach, which is capable of using censored data and producing quantitative output.
The new P-gp Efflux Ratio submodule allows you to:
- View predictions as quantitative Efflux Ratio (ER) values, characterizing the ratio of bidirectional transport rates in polarized transport assays
- View the underlying contributions of passive diffusion and active P-gp mediated efflux rates in the Caco-2 cell line at physiological conditions
- Edit text boxes which display the predicted values of key physicochemical descriptors used as inputs for the model
- Manually enter experimental values to increase prediction accuracy or simulate the effects of changing PhysChem properties on the compound’s efflux potential
- Use a heatmap to explore the necessary changes to the molecule’s PhysChem profile to achieve the desired transport characteristics
- Compare Efflux Ratios, binary classification, and the type of assay used for measurement with five most chemically similar structures from the training set
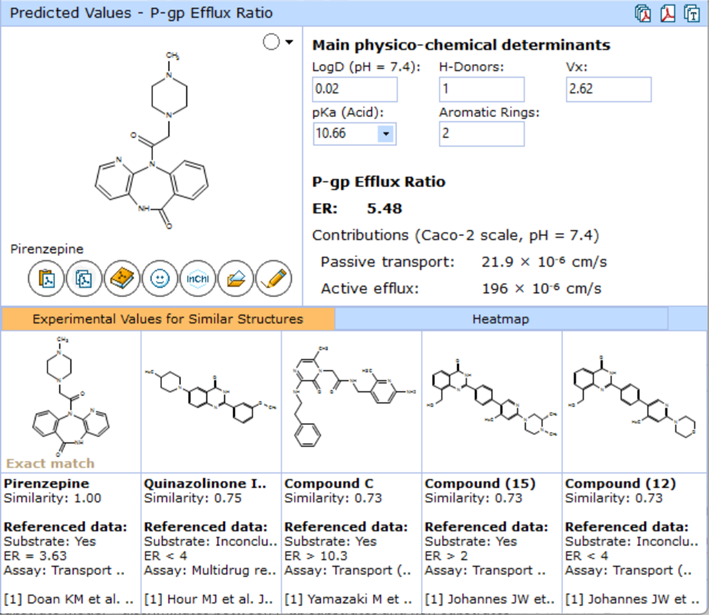
Predicted P-gp Efflux Ratio, also showing five most similar structures from the training set.

Heatmap indicates the position of the current compound (blue dot) in the physicochemical space of the training set.
Improved GALAS Model for P-gp Substrates
You can now expect improved accuracy of the GALAS model, including:
- Almost two-fold increase in the training library of the GALAS predictor to ~2900 compounds
- Inclusion of novel congeneric series from recently published lead optimization studies
- Increased applicability domain coverage for many new classes of drug-like chemicals
ACD/Labs’ development team is eager to collaborate with organizations to improve predictions for novel compounds. Do you have accurately measured experimental values for the predictions we support? Contact us to discuss how we may work together.
Want to learn more?
Read more about the full features of ADME Suite, or contact us for help upgrading your software.
